Epigenome modifications and intracellular signaling pathways play essential roles in regulating cellular phenotypes and disease processes, but they are often studied separately. Research in the EpiSignal RTG will focus on the interplay of these regulatory networks and the consequences of this crosstalk on normal cellular responses and the development of diseases.
For example, EpiSignal research projects will investigate how epigenome modifications affect cell signaling, how signaling pathways control chromatin regulation, and how these processes are coupled. The specific epigenetic mechanisms studied will include DNA and histone methylation, and the regulation of the activity of epigenetic enzymes.
Moreover, lysine and arginine methyltransferases active on histones but also on non-histone proteins will be investigated which directly connect epigenetic regulation with cell signaling networks. Within the cell signaling networks, EpiSignal will focus on kinase cascades, molecular scaffolds and protein posttranslational modifications part of the Tumor necrosis factor alpha (TNFα) and Transforming growth factor beta (TGFβ) pathways. The cellular outcomes of these molecular interplays, i.e. proliferation/arrest, survival/death, migration/invasion and differentiation/dedifferentiation will be studied in breast and colorectal cell systems of epithelial origin.
The EpiSignal research will make important contributions to the understanding of the regulation of cell differentiation and proliferation during normal development, shed new light onto the complex and interwoven networks and regulatory principles of cell and epigenome signaling systems, and provide novel insights into processes occurring during cancer onset and progression.
EpiSignal projects
DNA methyltransferases DNMT3A and DNMT3B play essential roles in development by regulating gene expression and chromatin organization. Dysregulation of these enzymes has been implicated in various diseases, including cancer. However, the mechanisms controlling DNMT3A/B enzymatic activity remain poorly understood. Post-translational modifications (PTMs) such as phosphorylation, ubiquitination, and arginine methylation are critical regulators of DNMT3A/B function, influencing their localization, stability, and activity. Despite their importance, the specific molecular effects of individual PTMs, as well as the enzymes and signaling pathways responsible for their modification, remain largely unknown. This project aims to identify and characterize PTMs that regulate DNMT3A/B activity and localization in the context of TGFβ-induced epithelial-mesenchymal transition (EMT) and breast cancer progression by integrating biochemical, proteomic, and functional assays.
Main supervisor: Prof. Dr. Angelika Hausser (Molecular Cell Biology)
Co-supervisor: Prof. Dr. Dirk Schwarzer
This project investigates the regulation and control of the DNA methyltransferase DNMT3A by interacting chromatin factors. DNMT3A’s enzymatic activity is regulated by post-translational modifications, interacting proteins, and histone modifications. Key interaction partners include P53, NF-κB, PU.1, MYC, MBD3, and epigenetic regulators such as H3K9 methyltransferases, histone deacetylases, and chromatin remodelers. This project aims to characterize the molecular interactions between DNMT3A and these factors using biochemical pull-down assays, cellular interaction and co-localization studies with fluorescence-tagged proteins. Peptide array binding experiments and mutagenesis will be conducted to map interaction interfaces precisely. Additionally, the effects of these interactions on DNMT3A’s activity and their role in targeted DNA methylation will be investigated in cells by whole-genome bisulfite sequencing. Ultimately, this research will enhance our understanding of DNMT3A’s role in epigenetic regulation and decipher how DNA methylation patterns are set up in cells. Furthermore, the study will explore the influence of DNMT3A and the DNMT3A interactors on dynamic DNA methylation patterns during TNFα and TGFβ signaling and thereby provide insights into epigenetic regulation downstream of these signaling pathways. Our findings will enlighten the interplay between DNA methylation and cellular signaling pathways, particularly in tumorigenesis and cancer progression.
Main supervisor: Prof. Dr. Albert Jeltsch (Biochemistry & Molecular Epigenetics)
Co-supervisor: Prof. Dr. Markus Morrison
This project explores the relationship between a novel RUNX2/PRMT6 epigenetic gene regulatory complex and TNFα signaling. The RUNX2 transcription factor is linked to various cancers, including colorectal cancer (CRC). Our findings indicate that RUNX2 interacts with the protein arginine methyltransferase 6 (PRMT6) at genes within the TNFα signaling pathway. This suggests that the RUNX2/PRMT6 complex regulates the expression of TNFα genes through PRMT6-mediated histone modifications. Consequently, this connection indicates an epigenetic role of the RUNX2/PRMT6 complex in TNFα signaling, potentially influencing cell proliferation and colorectal cancer progression. To further investigate this relationship, we aim to identify RUNX2/PRMT6 target genes associated with TNFα in colorectal cancer and examine the crosstalk between TNFα signaling, RUNX2, and PRMT6. These studies will provide valuable insights into the roles of RUNX2/PRMT6 and TNFα signaling in determining CRC cell fates, and uncover molecular targets for CRC therapy.
Main supervisor: Prof. Dr. Jörn Lausen (Molecular Genetics)
Co-supervisor: PD Dr. Philipp Rathert
TGFβ stimulation induces large-scale gene expression changes to initiate a wide range of cellular responses such as cell cycle arrest, cell migration, apoptosis. TGFβ-induced cellular decision making is perturbed in diseases such as cancer, but the underlying changes in signalling and gene-regulatory networks remain incompletely understood. By combining quantitative experiments, bioinformatics and modelling, this project will provide a systems-level description of TGFβ-induced chromatin modifications, gene regulation and decision making. Mathematical models will be trained using perturbation data at the signaling and gene expression levels to ultimately rewire cellular decision making in mammary epithelial cells and a panel of breast cancer cell lines.
Main supervisor: Prof. Dr. Stefan Legewie (Systems Biology & Biochemistry)
Co-supervisor: Prof. Dr. Nicole Radde
This project studies how 5AD-induced epigenome changes affect the TNFα/TNFR1 interactome as a whole and how cell survival vs. death responses will be affected in colorectal cancer cell models. It will also investigate if and how methylation patterns of prominent promoter regions of TNFα pathway regulators can be linked to 5AD-induced changes or restoration of protein amounts, and if these could therefore carry potential as predictors for regained responsiveness. Likewise, it will unravel how robust 5AD-induced epigenetic changes are against cellular efforts to restore prior epigenetic states through intrinsic DNA methylation, which could give rise to regaining treatment resistance.
Main supervisor: Prof. Dr. Markus Morrison (Cell Biology & Systems Biology)
Co-supervisor: Jun. Prof. Dr. Franziska Traube
This project will explore how Deleted in Liver Cancer 1 (DLC1), a Rho regulator and key tumor suppressor, influences not just cytoskeletal dynamics but also nuclear processes linked to gene regulation. Our recent findings suggest that DLC1 interacts with chromatin factors, hinting at a previously unrecognized role in epigenetic control. Here we will explore how Rho signaling and DLC1 shape epigenetic programs during TGFβ-driven epithelial-mesenchymal transition (EMT) - a key process in cancer invasion and metastasis. Using cutting-edge approaches, including genetically encoded fluorescent reporters to visualize epigenetic activity in living cells, global transcriptomic profiling, and advanced 2D/3D cancer models, we will: (i) Investigate how Rho signaling and DLC1 influence the transcriptional landscape during EMT. (ii) Identify key epigenetic proteins that modulate DLC1 function. (iii) Determine how these interactions drive cancer cell differentiation and transformation. This interdisciplinary project offers the chance to work at the intersection of cancer biology, epigenetics, and cell signaling, providing novel insights into tumor progression and potential therapeutic avenues.
Main supervisor: Prof. Dr. Monilola Olayioye (Molecular Tumor Cell Biology)
Co-supervisor: Prof. Dr. Jörn Lausen
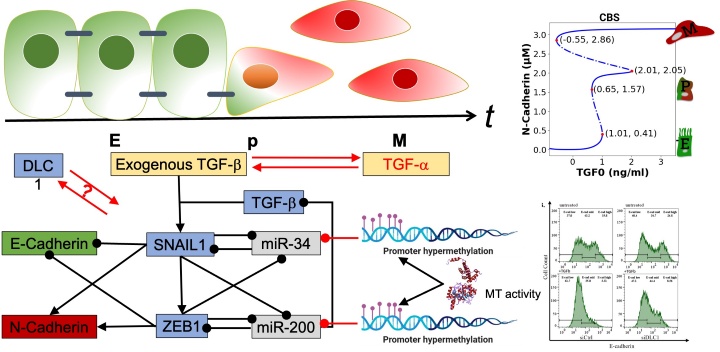
Epithelial-to-mesenchymal transition (EMT) is a physiological process in which epithelial cells are converted to a mesenchymal phenotype, a process that is frequently hijacked in cancer. It is now known that EMT occurs in a stepwise manner, involving metastable partial EMT (pEMT) states. Due to their plasticity and reversibility, these pEMT states are considered particularly relevant for the survival of cancer cells and the development of metastases. By combining mathematical modeling, statistical methods for model calibration to experimental data, and nonlinear dynamics methods, in this project we will explore the dynamics of EMT and MET and gain insights into the modulation of pEMT states by intra- and extracellular signaling crosstalk. Specifically, we will investigate perturbations and molecular crosstalk at three different levels: alterations of the core network through epigenetic regulation, interactions of the core network with molecular factors deregulated in cancer, and crosstalk effects of TGFß with TNFα signaling. Results will deepen our mechanistic understanding about how intracellular and extracellular cues modulate EMT dynamics in non-transformed breast epithelial cells and, as a future perspective, how model hypotheses can be translated to breast cancer cells defining their differentiation states.
Main supervisor: Prof. Dr. Nicole Radde (Modeling & Simulation of Cellular Systems)
Co-supervisor: Prof. Dr. Monilola Olayioye
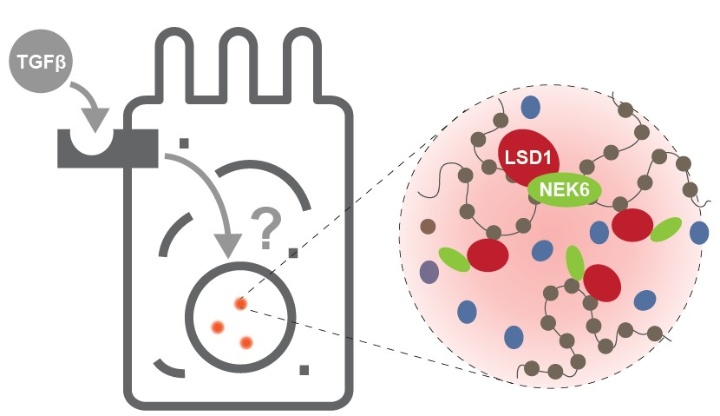
Phase separation is emerging as a fundamental principle in cellular organization, allowing cells to dynamically compartmentalize molecular components without the need for membranes. In the nucleus, liquid-liquid phase separation (LLPS) plays a crucial role in structuring chromatin environments and organizing transcriptional machinery. This project delves into how NEK6 and LSD1 form chromatin sub-compartments (CSCs) through phase separation and how these structures impact TGFβ signaling, a pathway vital for cell fate decisions, differentiation, and cancer progression. By applying advanced imaging, transcriptomics, and functional assays, we aim to uncover how these condensates shape the transcriptional landscape and whether they act as regulatory hubs for TGFβ-induced gene expression. This research not only advances our understanding of LLPS in gene regulation but may also reveal novel therapeutic opportunities targeting epigenetic condensates in cancer and other diseases. Through biophysical, transcriptomic, and functional analyses, we will determine the impact of these condensates on gene expression and cellular responses, particularly in the context of cancer progression.
Main supervisor: PD Dr. Philipp Rathert (Biochemistry & Molecular Epigenetics)
Co-supervisor: Prof. Dr. Stefan Legewie
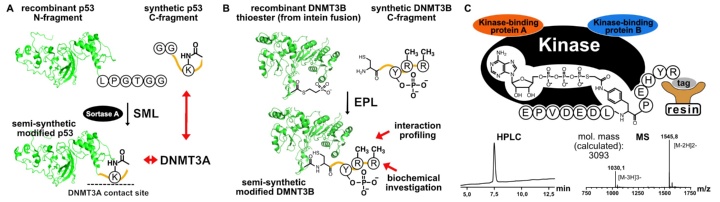
DNA methyltransferases display complex protein-protein interaction networks that impact their roles in regulating gene expression. These interaction networks are modulated by signalling-induced posttranslational modification (PTM) patterns, which can hardly be addressed by conventional methods of biochemistry. We plan to use protein semi-synthesis based on sortase-mediated ligation (SML) and expressed protein ligation (EPL) to generate variants of DNA methyltransferases DMNT3A and DMNT3B with defined PTMs and investigate the impact of the introduced modifications on signalling and epigenetics. In addition, tailor-made probes for capturing kinases that phosphorylate DNMT1 will be established and used for proteomics-profiling of the kinase complexes that regulate this enzyme. This project provides an excellent training opportunity for young investigators interested in chemical protein engineering for academic and industrial applications.
Main supervisor: Prof. Dr. Dirk Schwarzer (Chemical Biology & Biochemistry)
Co-supervisor: Prof. Dr. Albert Jeltsch
Ten-eleven translocation (TET) enzymes are a protein family of α-ketoglutarate-dependent dioxygenases that oxidize the most prevalent epigenetic nucleobase 5-methylcytosine to 5-hydroxymethylcytosine and further to 5-formyl- and 5-carboxycytosine. The latter two can be subsequently removed by DNA-glycosylases and replaced by unmodified cytosine, resulting in DNA demethylation. TET-deficiency is a well-known driver of tumorigenesis, but direct links between TET activity and phenotypic outcomes are largely unexplored. The aim of this project is to elucidate on the mechanistic level how TET2-function is linked to tumorigenesis and TGFβ-induced EMT in colon carcinoma by investigating in a holistic manner the TET2-interactome. To elucidate TET2-protein interaction partners we will use mass spectrometry-based proteomics and identify in the subsequent computational analysis functional protein complexes that link epigenetic signalling to other cellular signalling pathways.
Main supervisor: Jun. Prof. Dr. Franziska Traube (Biochemistry of Cellular Biomedical Systems)
Co-supervisor: Prof. Dr. Angelika Hausser
Contact Us

General contact email
episignal@ibc.uni-stuttgart.de

Albert Jeltsch
Prof. Dr.Acting Director Institute of Biochemistry, Speaker EpiSignal RTG

Sara Weirich
Dr.Group leader Institute of Biochemistry, Program manager EpiSignal RTG










